Data quality control procedures, 15 August 2021
GISAID EpiCoV™ data quality control procedures and B.1.621 (Mu) monitoring
In late July 2021, GISAID confirmed with public health authorities its procedures for the quality control of gaps in genomic sequences that cause frameshifts and other genomic changes. Social media discussions mistakenly suggested B.1.621 (Mu) genome sequences could have been kept from release due to a 4nt deletion in ORF3A that results in a premature stop codon.
Insertions and deletions in genome sequences may result in frameshifts, which in turn can result in truncated non-functional proteins that are normally selected against during natural selection and hence more likely a technical error of genome sequencing techniques. To account for different scenarios, when frameshifts are detected, GISAID protocols call for curators to confirm with data submitters that the observations made are correct and not resulting from bioinformatics errors. As a result of GISAID’s confirmation procedures, countless errors are detected that would otherwise be introduced in the EpiCoV™ database.
With at least 18-months of evolution, some of these viral sequence changes begin to occur naturally. Several of these changes have already been confirmed by submitters. By mid-August 2021, over 100,000 frameshifts have been confirmed and are noted in the data records, the earliest of which was detected and confirmed in April 2020 (EPI_ISL_419444). Many advanced sequencing laboratories, in particular those generating high volumes of data (e.g. COG-UK), have already tuned their bioinformatics pipelines to detect insertions and deletions more reliably, while others are building capacity in this area. Since late 2020, these changes passing their pipelines are directly accepted in GISAID’s EpiCoV™ curation workflow.
Consequently, the vast majority of sequences with credible changes are made available in the EpiCoV™ database without the need for additional confirmation by submitters. Social media discussions suggesting that B.1.621 (Mu) genome sequences with any such changes would remain undetected, or would delay their release were unwarranted.
GISAID data curation protocols do not permit the rejection of sequences. GISAID’s inclusion criteria only call for genome sequences to have a length over 100 nucleotides; and less than 50% stretches of Ns.
For those submission where frameshifts cannot be confirmed by the submitter, the genome sequences are tagged with the comment “frameshift not confirmed”.
By mid-August, the relative frequency of B.1.621 (Mu) cases observed remained low (Fig. 1) when compared to other variants of hCoV-19. For example, the variant detected with highest frequency during the same time period was the B.1.617.2/AY.* (Delta) variant with 60-fold higher submissions.
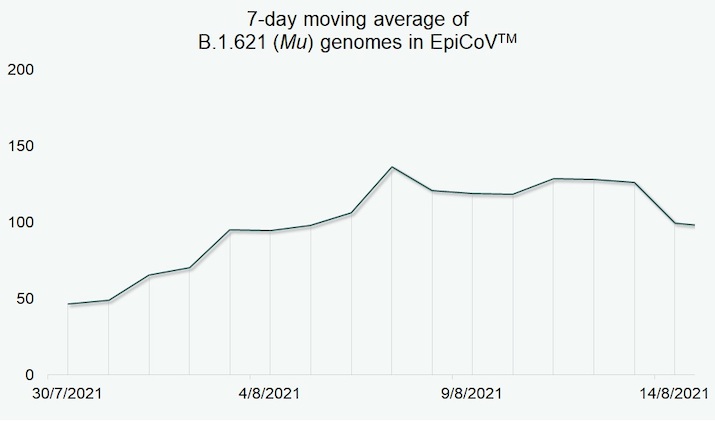
Figure 1. B.1.621 (Mu) submissions in EpiCoV™ between 30 July and 14 August 2021
The small fraction of entries released with frameshift mutations continued stable over the same time period (Fig. 2), indicating that GISAID’s frameshift handling protocols did not affect the availability of B.1.621 (Mu) genomes in EpiCoV™.
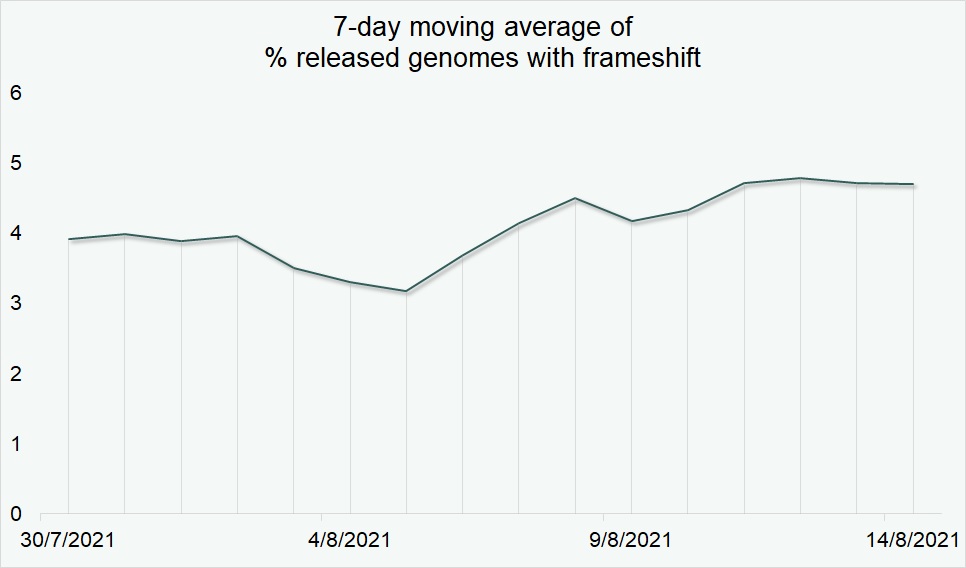
Figure 2. Stable release pattern of entries with frameshifts normalized with respect to entries without frameshifts
The GISAID Emerging Variants tracker was deployed in February 2021, and enables automated monitoring and ranking of new combinations of relevant spike protein changes in EpiCoV™ that may become variants of interest (VOI) or variants of concern (VOC) in the future. B.1.621 appeared ranked in the top 5 variants in the Emerging Variants tracker (Fig. 3) as early as June 2021.
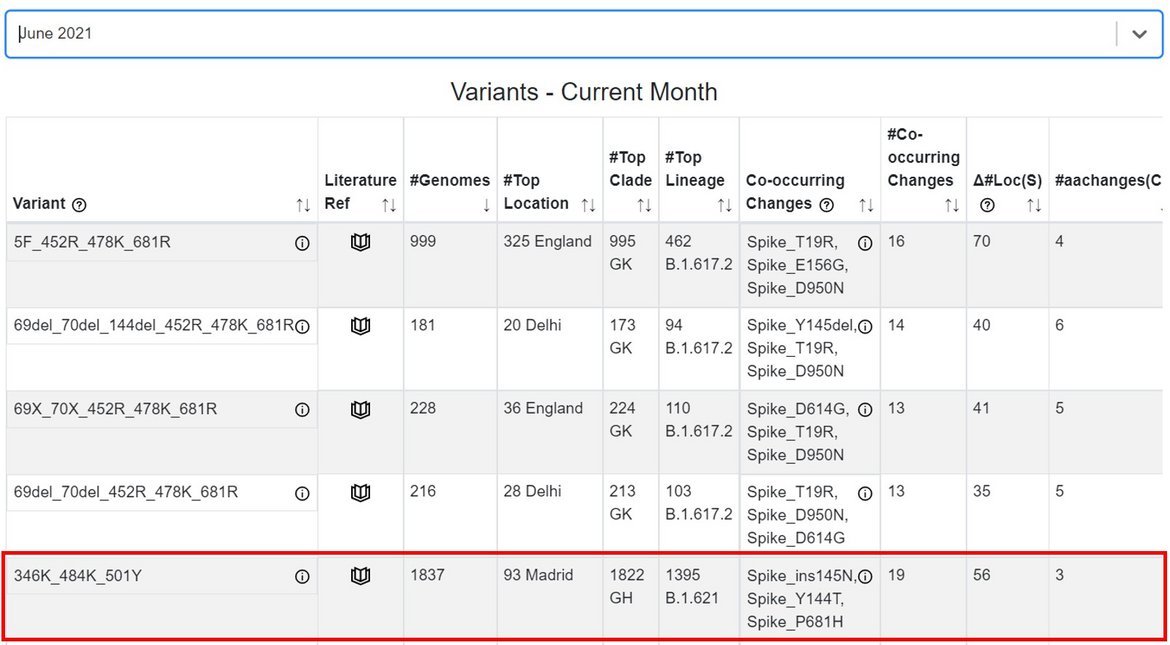
Figure 3. EpiCoV™ Emerging Variants tracking for hCoV-19 genomes with collection date in June 2021
GISAID data and tools thus enable public health authorities such as the World Health Organization and other partners to monitor and respond to viral genomic changes in a timely manner based on scientific evidence.

