Comment on recent variants and spike protein changes
As seen on many occasions before, mutations are naturally expected for viruses and are most often simply neutral regional markers useful for contact tracing. The changes seen have rarely affected viral fitness and almost never affected clinical outcome but the detailed effects of these mutations remain to be determined fully. Changes in the spike protein have relevance for potential effects on both host receptor as well as antibody binding with possible consequences for infectivity, transmission potential and antibody and vaccine escape. Actual effects need to be measured and verified experimentally.
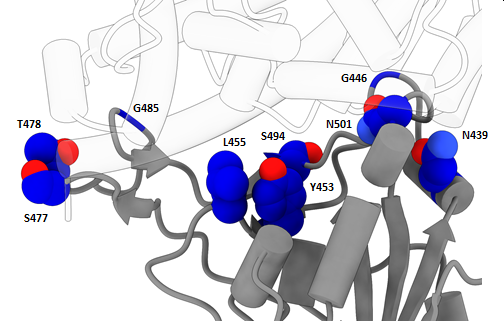
GISAID reports updates on spike mutations of recent submissions via gisaid.org/spike and any sequence can be tested for spike mutations via gisaid.org/covsurver and from the internal analysis interface where individual countries/regions and time periods can be selected for custom analysis. This allows highlighting and tracking the rise of mutations like D614G or the currently most common receptor binding mutations S477N (part of large Melbourne outbreak from clade GR and some Central European clade GH clusters), N439K (long lasting UK outbreak with clade G and European spillover), N501Y (part of new UK variant VUI-202012/01 in clade GR as well as a recent clade GH outbreak in South Africa) and Y453F (mink adaptation and part of limited outbreaks) as well as combinations of these mutations with deletions altering the spike protein surface.
As has become evident, these few S gene mutations and some deletions are found in multiple genomic contexts (different clades in different countries) that may be an early indication for some potential advantage for these viruses but needs to be verified and does not necessarily mean change in clinical severity or transmission efficiency.
GISAID is tracking occurrence and spread of commonly discussed variants at gisaid.org/variants